 |
Gene Ontology Annotation Tool (GOAT) |
 |
Gene Ontology Annotation Tool (GOAT) |
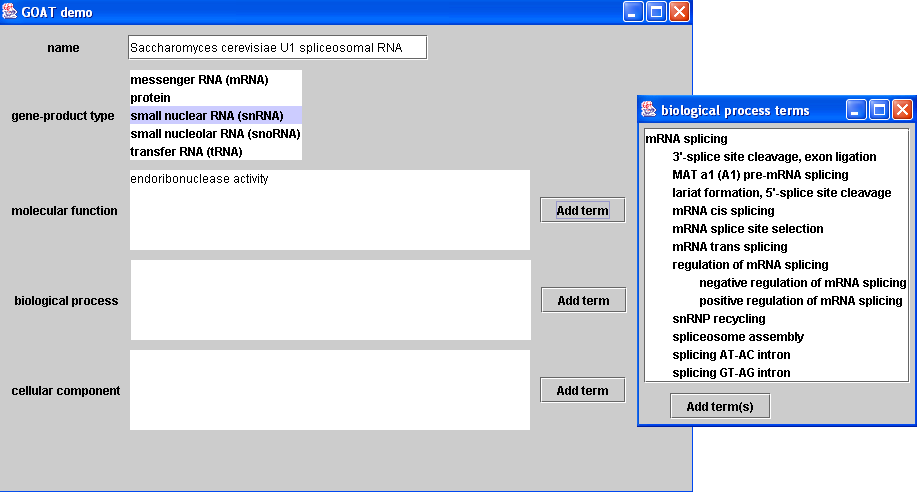
This is a screenshot of GOAT as currently implemented. In the main window (on the left), it can be seen that each gene-product annotation can include a free-text name, a gene-product type selected from an enumerated list, and a set of one or more GO terms for each of the three GO subontologies. It is in these GO-subontology fields where users can be aided in the process of annotation. For example, as shown in this figure, the user has entered “endoribonuclease activity” in the molecular-function field and has chosen “small nuclear RNA (snRNA)” from the menu of gene-product types. Upon indicating that she wishes to enter a value for the biological-process field (by pressing the corresponding "Add term(s)" button), GOAT will dynamically attempt to retrieve the association record for “endoribonuclease activity” from the instance store by forming and submitting DIG queries. It also asks the reasoner directly for GO-term concepts that are explicitly associated with “small nuclear RNA” in the loaded DAML+OIL version of GO. A set of biological-process terms found to be associated with all of this information is determined, and these terms’ descendants are dynamically retrieved from the ontology. This subset of GO terms is shown as a tree (in the familiar GO format rather than that of our DAML+OIL version) in a pop-up window (on the right), from which she may choose one or more terms as field values. Thus, instead of searching for the appropriate term(s) through all of GO, the user is presented only with values likely to be appropriate.
As can be discerned, GOAT is still a prototype rather than a complete annotation tool.
Contact Mike Bada or Robert Stevens for more information.
Last modified 4 April 2004 by Mike Bada.